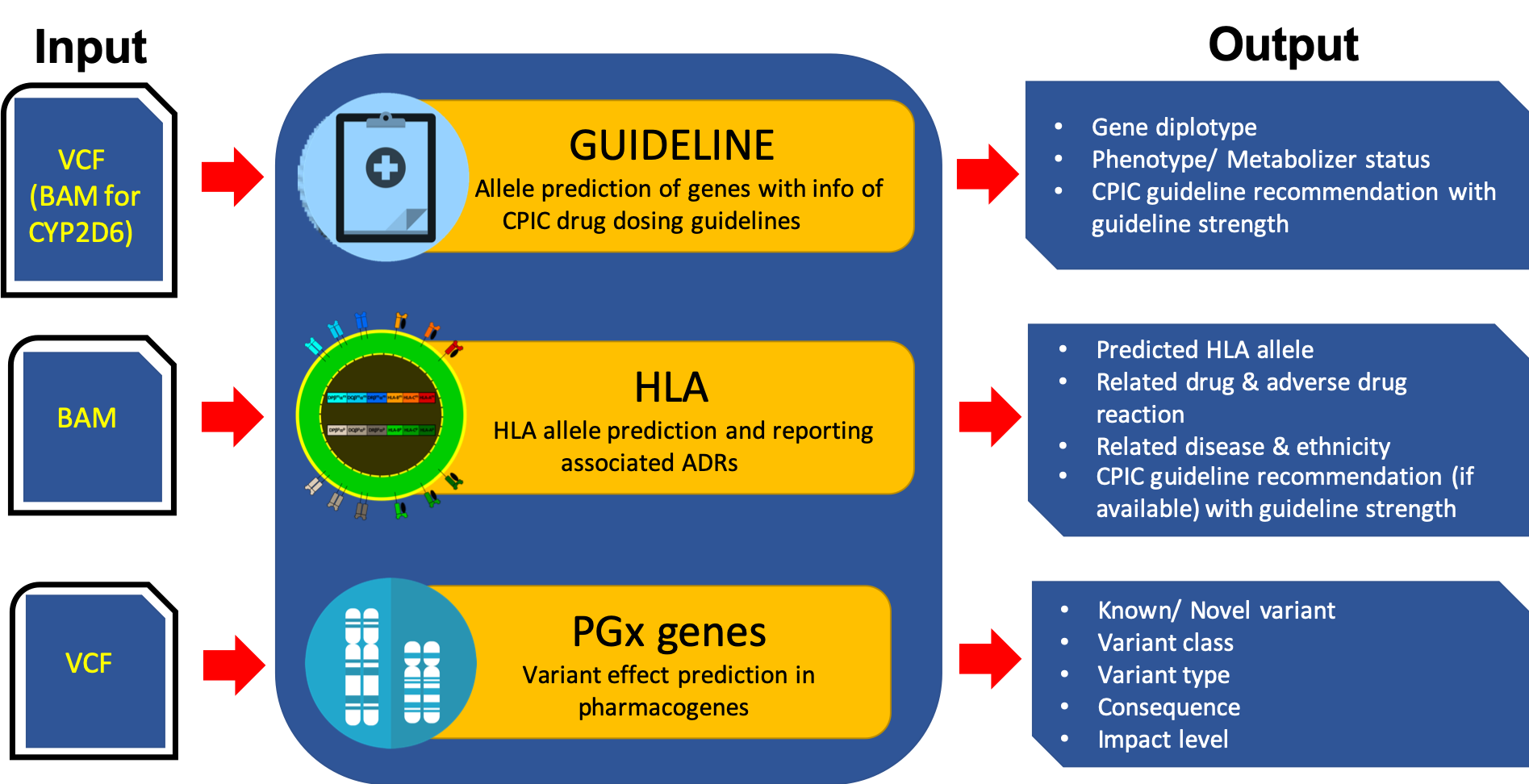
Overview of PharmVIP analysis modules
The current release of PharmVIP (beta version) consists of three analysis modules.
1) Guideline module:
Function: Predict the haplotypes/alleles in 17 genes which are associated with 47 drugs and have information of CPIC (Clinical Pharmacogenetics Implementation Consortium) drug dosing guidelines.
Required Input: VCF/gVCF file (CYP2D6 analysis requires BAM file)
Features: User can choose to analyze from the gene list or the drug list (grouped by therapeutic category). Allele matching and dosing guideline assignment are performed by our own algorithm/method. CYP2D6 allele is predicted by Astrolabe software. The output can be filtered by selecting the displayed genes, phenotypes, and guideline strengths.
Output: Predicted diplotype, metabolizer status/phenotype, CPIC dosing recommendation with guideline strength
Resources:
- CPIC guideline recommendations: PharmGKB (https://www.pharmgkb.org/) (downloaded on 20-02-2020)
- Allele definition tables: PharmGKB (https://www.pharmgkb.org/) (downloaded on 20-02-2020)
2) HLA module:
Function: Predict the alleles in all HLA genes and report the published adverse drug reactions (ADRs) associated with these HLA alleles.
Required Input: BAM file
Features: User can choose the cohort ethnicities of the reported ADR cases in the HLA-ADR association studies. The output can be filtered by selecting the displayed HLA genes, drugs, ADRs, cohort ethnicities, and diseases of patients with ADRs.
Output: Predicted HLA alleles, the associated drugs and adverse drug reactions, patient diseases and ethnicities, CPIC guideline recommendations (if available) with guideline strength
Resources:
- Known adverse drug reactions with HLA alleles: The HLA and Adverse Drug Reaction Database (http://allelefrequencies.net/hla-adr) (downloaded on 20-02-2020)
- HLA sequence library: The IPD-IMGT/HLA Database (https://www.ebi.ac.uk/ipd/imgt/hla) (Release 3.35.0)
3) PGx genes module:
Function: Identify genomic variants (single nucleotide polymorphisms, insertions, deletions) in 3,533 pharmacogenes listed from different resources and predict their effects to gene function.
Required Input: VCF/gVCF file
Feature: User can choose a gene list to be analyzed. The output can be filtered by variant classes, types, consequences, impacts, and SIFT/PolyPhen classes
Output: List of known/novel variants, variant types, variant classes, consequences, impact level
Resources:
- The list of pharmacogenes from:
- CPIC guideline : https://www.pharmgkb.org/guidelineAnnotations (downloaded on 20-02-2020)
- DrugBank : https://www.drugbank.ca/releases/latest#protein-identifiers (version 5.1.5)
- FDA PGx Biomarkers : https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling (downloaded on 20-02-2020)
- iPLEX PGx Pro Panel : https://agenabio.com/wp-content/uploads/2016/03/51-20037R3.0_iPLEX-PGx-Pro-Panel-Flyer_HIGH-WEB.pdf
- Korean Target seq (Han 2017) : Han SM, Park J, Lee JH, Lee SS, Kim H, Han H, Kim Y, Yi S, Cho JY, Jang IJ, Lee MG. Targeted Next-Generation Sequencing for Comprehensive Genetic Profiling of Pharmacogenes. Clin Pharmacol Ther. 2017; 101(3): 396-405. doi: 10.1002/cpt.532. (PMID: 27727443)
- PGRNseq (Gordon 2016) : Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics. 2016; 26(4): 161-168. doi: 10.1097/FPC.0000000000000202. (PMID: 26736087)
- PGx testing (Gamazon 2012) : Gamazon ER, Skol AD, Perera MA. The limits of genome-wide methods for pharmacogenomic testing. Pharmacogenet Genomics. 2012; 22(4): 261-72. doi: 10.1097/FPC.0b013e328350ca5f. (PMID: 22344246)
- PharmGKB clinical variant data : https://www.pharmgkb.org/downloads (downloaded on 5-02-2020)
- PharmGKB VIPs : https://www.pharmgkb.org/vips (downloaded on 20-02-2020)
- TruGenome Illumina : https://www.illumina.com/content/dam/illumina-marketing/documents/clinical/trugenome-intended-use-pharmacogenomics-screen.pdf (downloaded on 15-04-2019)
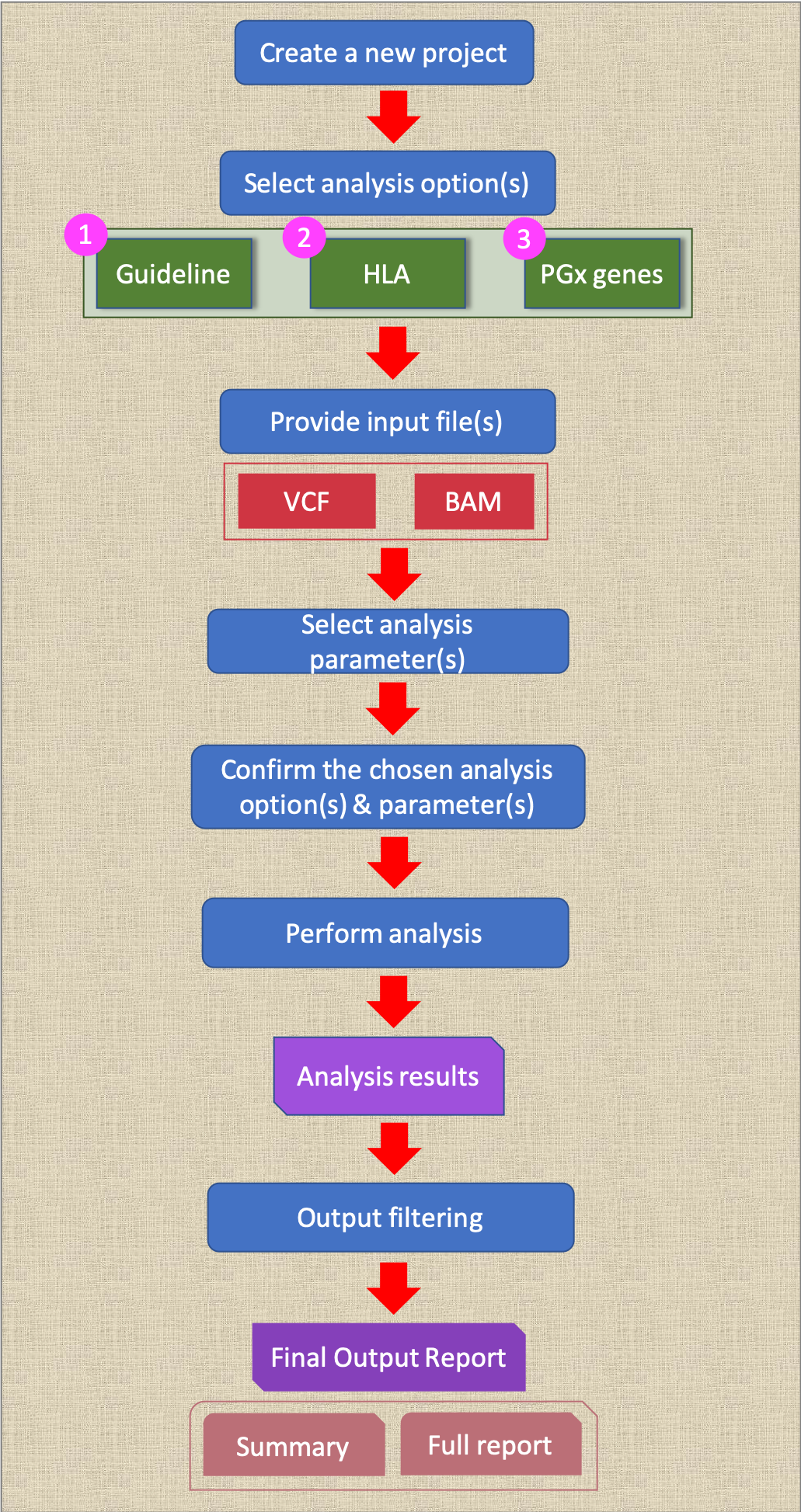
Summary of user’s analysis workflow



